Precision Medicine to Drug Discovery
Transform patient DNA into therapeutic molecule candidates in under 5 hours.
GPU-accelerated genomics, AI-powered reasoning, and generative drug discovery.
What It Is
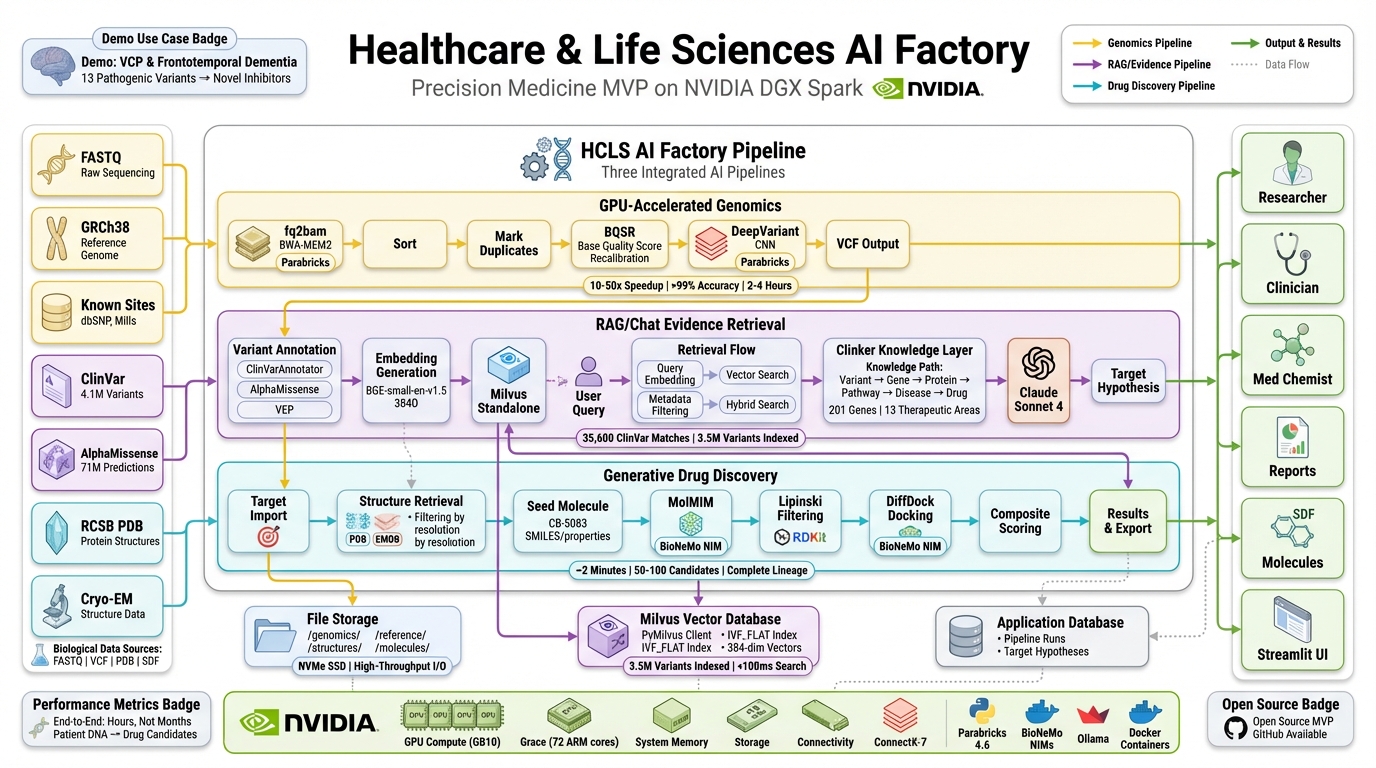
The Healthcare & Life Sciences (HCLS) AI Factory unifies three production-grade AI workflows into a single, continuous system — designed to take raw patient DNA and produce viable drug candidates without the fragmentation and delays that define traditional approaches.
This enables computational predictions. They're promising starting points for laboratory testing, not finished medicines. Real drug development requires years of laboratory and clinical validation. But what this HCLS AI Factory does is collapse the first and most expensive step — identifying targets and generating promising candidates — from months of work into hours.
Raw FASTQ files flow through NVIDIA Parabricks for GPU-accelerated genomics — alignment, variant calling, clinical-grade accuracy via DeepVariant — completing in hours instead of days.
Outputs feed directly into an evidence layer where millions of variants can be queried in natural language, grounded in ClinVar, AlphaMissense, structural data, and curated biomedical knowledge.
Validated targets then move into generative drug discovery via NVIDIA BioNeMo, where novel molecules are created, docked, scored, and ranked.
No batch jobs. No manual handoffs. Full lineage from patient DNA to candidate therapeutic.
One workstation. One workflow. Hours, not months.
Three-Stage AI Pipeline
End-to-end from raw sequencing data to drug candidates
GPU Genomics
200 GB FASTQ processed via NVIDIA Parabricks and DeepVariant. 11.7M variants called at >99% accuracy.
120 – 240 minExplore pipeline →
Evidence RAG
3.5M variants annotated, embedded, and indexed. AI-powered reasoning identifies 171 druggable targets across 13 therapeutic areas.
InteractiveExplore pipeline →
Drug Discovery
BioNeMo MolMIM generates novel molecules. DiffDock predicts binding. RDKit scores drug-likeness. 100 ranked candidates per target.
8 – 16 minExplore pipeline →
By the Numbers
Real results from a single NVIDIA DGX Spark workstation
Traditional vs. HCLS AI Factory
What used to take months now takes hours
| Metric | Traditional Approach | HCLS AI Factory |
|---|---|---|
| Sequence Alignment | 12 – 24 hours | 2 – 3 hours |
| Variant Calling | 8 – 12 hours | 1 – 2 hours |
| Annotation & Interpretation | Days of manual work | Minutes (automated) |
| Target Identification | Weeks of literature review | Minutes (AI-powered) |
| Drug Candidate Design | Months of medicinal chemistry | 8 – 16 minutes |
| Total Time | Weeks to months | < 5 hours |
| Infrastructure Cost | $100K+ (cluster) | $3,999 (DGX Spark) |
| Reproducibility | Variable | Deterministic |
Origin
In 2012, I set out to use my high-performance computing background for something that mattered. I started with one conviction: no parent should ever have to lose a child to disease.
That conviction led me to Pediatric Neuroblastoma. I taught myself biology, genomics, molecular pathways, drug discovery — whatever the work required. I made one commitment early: I would not profit from this. Whatever I built, I would give away freely, so others could build on it and move faster than any one person ever could alone.
Thousands of hours later, this is the result.
— Adam Jones
Why This Is Open
This project is open by design, not as a shortcut or a visibility exercise, but as a deliberate decision about how foundational healthcare infrastructure should be built.
The challenges in precision medicine are no longer primarily scientific — they are architectural. Fragmented pipelines, opaque tooling, and closed systems slow the transition from genomic data to actionable insight.
By open-sourcing the HCLS AI Factory, this project provides a reproducible, inspectable reference implementation for end-to-end genomics, AI reasoning, and therapeutic exploration.
Open access to infrastructure knowledge accelerates progress, enables collaboration, and shifts innovation away from re-solving plumbing toward advancing care.
Who This Is For
For people and institutions working at the intersection of healthcare, life sciences, and AI who need systems, not abstractions.
Researchers & Bioinformaticians
Building, extending, or validating secondary genomics pipelines and variant interpretation workflows who need reproducible, inspectable infrastructure.
Clinicians & Translational Teams
Exploring how genomic data, AI reasoning, and therapeutic insights can be integrated into real-world decision-making.
Academic Medical Centers
Teaching, researching, or operationalizing genomics and AI at scale, where transparency, reproducibility, and extensibility are critical.
AI & Systems Engineers
Interested in how real biomedical workloads behave when treated as first-class AI systems, including data flow, orchestration, and reasoning layers.
Platform Builders
Designing future healthcare platforms who want a concrete reference architecture for AI-native pipelines rather than high-level diagrams.
Anyone, Anywhere
This project is not limited to a single specialty, disease, or institution. It is designed to be a shared foundation that can support many domains.
What This Is Not
The intent is clarity, not control.
It is a reference architecture and research platform, not a regulated medical device or diagnostic system.
All workflows, data flows, and reasoning layers are inspectable and reproducible.
It is designed to augment researchers and clinicians, not automate judgment.
The system is modular by design and intended to be adapted, extended, or specialized.
The architecture is deliberately vendor-neutral and infrastructure-agnostic.
Technology Stack
Take it. Use it. Make it better.
Everything is open source under Apache 2.0. Deploy the full pipeline on your DGX Spark in minutes.